Research.The long-term goal of our research is to understand how the actin cytoskeleton regulates cell migration and adhesion in an in vivo context. We use the power of C.elegans genetics and imaging to investigate the role of actin regulators during cell migration in vivo.
|
Research
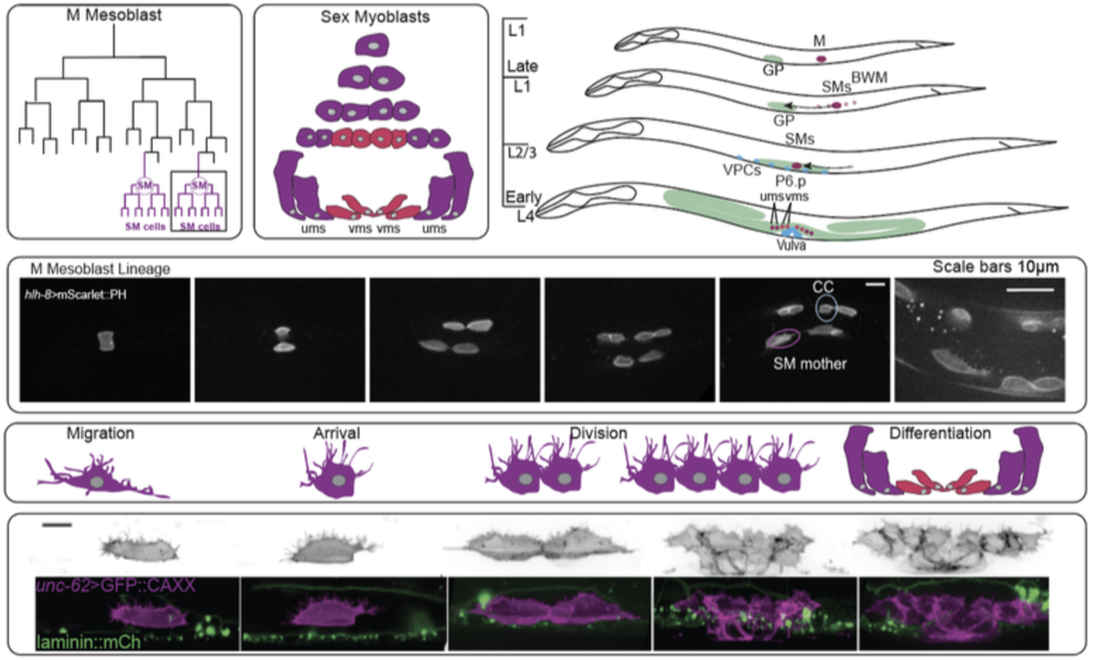
We use the sex myoblast cells of C.elegans to probe the role of the actin cytoskeleton in long distance cell migration. The sex myoblasts (SMs) migrate ~65 microns during the second larval stage to arrive at the center of the gonad where they then divide and differentiate into the uterine and vulval muscle cells (ums and vms). We have established protocols to image this migration and are currently working on quantification of membrane and actin dynamics. Preliminary data, that the lab will confirm and expand upon, shows that key actin regulators are required for this process. Future research endeavors will address if this mechanism of actin based migration is conserved in the presomitic mesodermal cells of zebrafish.
|
Check out what we worked on last summer!CollaborationsTe-Wen Lo at Ithaca College: Elucidating the role(s) of FGFR in the chemoattaracion of the migratory sex myoblasts in C. elegans
Emily Mace at Columbia University Irving Medical Center: Uncovering the migratory mechanisms of natural killer cells in complex 3D microenvironments Abhishek Kumar at the Marine Biological Laboratories, Woods Hole, MA: Using novel imaging platforms and modalities to image complex biological processes. The Adikes lab is looking forward to collaborating with scientists who design and use advanced imaging techniques to visualize highly dynamic cellular and sub-cellular events! MentoringThe Adikes lab fosters the growth and technical training of undergraduate scholars in a supportive and engaging environment!
|
PublicationsMondal, C., Gacha-Garay, M.J., Adikes R.C., Larkin, K., Di Martino J, Chien C-C., Fraser, M., Eni-aganga, I., Agullo-Pascual, E., Ozbek,U., Naba, A., Gaitas, A., Fu, T-M., Upadhyayula, S., Betzig, E., Matus, D.Q., Martin, B.L., and Bravo-Cordero, J.J. (2022) A proliferative-to-invasive switch is mediated by srGAP1 downregulation through the activation of TGFβ2 signaling. Cell Reports.
Palmisano, N.J.#, Azmi, M.A.#, Medwig-Kinney, T.N., Moore, F.E.Q., Rahman R., Zhang, W, Adikes, R.C.%, Matus, D.Q.%. (2022). A laboratory module that explores RNA interference and codon optimization through fluorescence microscopy using Caenorhabditis elegans. CourseSource DOI:https://doi.org/10.24918/cs.2022.15 #these authors contributed equally, %co-corresponding Morabito, R. D., Adikes, R. C., Matus, D. Q., Martin, B. L. (2021) Cyclin-Dependent Kinase Sensor Transgenic Zebrafish Lines for Improved Cell Cycle State Visualization in Live Animals. Zebrafish. DOI: 10.1089/zeb.2021.0059 Smith, J.J., Xiao, Y., Medwig-Kinney, T.N., Martinez, M.A.Q., Moore, F.E.Q., Palmisano, N.J., Kohrman, A.Q., Chandok Delos Reyes, M., Adikes, R.C., Liu, S., Bracht, S.A., Zhang, W., Wen, K., Krastsios, P., Matus, D.Q. (2021) The SWI/SNF chromatin remodeling assemblies BAF and PBAF differentially regulate cell cycle exit and cellular invasion in vivo. PLOS Genetics DOI:http://doi.org/10.1371/journal.pgen.1009981 Adikes, R.C.#, Kohrman, A.Q.#, Martinez, M.A.Q.#, Palmisano, N.J., Smith, J.J., Medwig-Kinney, T.N., Min M., Sallee, M.D., Ahmed, O.B.*^, Kim, N.^, Liu, S., Morabito, R.D., Weeks, N., Zhao, Q., Zhang, W., Feldman, J.L., Barkoulas, M., Pani, A.M., Spencer S.L., Martin, B.L., and Matus, D.Q. (2020). Visualizing the metazoan proliferation-differentiation decision in vivo. eLife 9:e63265 DOI: 10.7554/eLife.63265 #these authors contributed equally |
Invited TalksAdikes, R.C. (2023) The path to the light sheet: consideration for sample preparation. Light Sheet Fluorescence Microscopy Workshop. Marine Biological Laboratories, Woods Hole, MA
Shehzad, S.A. and Fitian, M.A. (2023) Investigating roles of cytoskeletal genes in C. elegans sex myoblast development. Capital District Imaging Seminar Series. Virtual Adikes, R.C. (2022) Roles of cytoskeletal genes in C.elegans sex myoblast development. RPI, Troy, NY. Adikes, R.C. (2022) How cells move and navigate complex environments. Bennington College Science Workshop, Bennington, VT Adikes, R.C. (2022) Matching Sample to System. Light Sheet Fluorescence Microscopy Workshop. Marine Biological Laboratories, Woods Hole, MA Adikes, R.C. (2021) Membrane and cytoskeletal dynamics during muscle progenitor migration. ASCB Cell Bio Annual Meeting subgroup on Cytoskeletal dynamics in health and disease. Virtual
Adikes, R.C. (2021) Regulation of cell migration during development and disease. Siena College Biology Department Seminar. Virtual |
Poster Presentations
|
Students Maria Fitian (left) and Bekkah Clauson (right) present their posters at the Spring Academic Showcase at Siena College.
Left to right: Maria Fitian, Sana Shehzad, Marty Chalfie (Nobel Prize for the discovery of GFP and a C. elegans researcher) and Mishal Razi in front their poster titled: Roles of Cytoskeletal Genes in C.elegans Sex Myoblast Development at the 2022 American Society for Cell Biology Annual Meeting.
Sana Shehzad presents her poster at the Spring Academic Showcase at Siena College.
|
Shehzad, S.A.*, Fitian, M.A.*, Razi M.*, Philip A., Adikes, R.C. The role of actin-binding proteins in cell migration during development. Spring 2023 Siena Academic Showcase.
Razi M., Fitian, M.A., Philip A., Shehzad, S.A., Adikes, R.C. The role of focal adhesions in SM cell migration. Spring 2023 Siena Academic Showcase Fitian, M.A., Fitian, M.A., Razi M., Shehzad, S.A., Adikes, R.C. Investigating the role of ring-forming proteins in cell migration. Spring 2023 Siena Academic Showcase. Philip A., Shehzad, S.A., Fitian, M.A., Razi M., Adikes, R.C. The effect of signaling proteins on cell migration during development of C.elegans. Spring 2023 Siena Academic Showcase Clauson, R., Fang, A., Ahmed O.B., Martinez M.A.Q., Matus, D.Q., Stern, M., Adikes, R.C. Examining the relationship between cell cycle and cell migration rates. Spring 2023 Siena Academic Showcase. Razi, M., Fitian, M., Shezhad, S., McKnight, T. Ahmed O.B., Matus, D.Q., Gibney, T., Pani, A.P., Adikes, R.C. Roles of Cytoskeletal Genes in C.elegans Sex Myoblast Development. 2022 ASCB Annual Meeting Fitian, M.A., Razi M., Shehzad, S.A. McKnight, T., Adikes, R.C. Investigating the Roles of Cytoskeletal Genes in SM Cell Migration in C. elegans. 2022 DBNY Undergraduate Research Symposium, Ithaca College. Maria won a poster prize! Clauson, R., Sterling, B., Matus, D.Q., Martin, B.L., Deng, Y., Adikes, R.C., Computation pipline to analyze protrusion dynamics. 2022 DBNY undergraduate research symposium, Ithaca College. Shezhad, S., McKnight, T., Adikes, R.C. Cell migration in C. elegans as a model for development and disease. 2022 Siena Academic Showcase Adikes, R.C., Moore, F.E.Q, Fang, A., Martinez M.A.Q.M., Ahmed O.B., Medwig-Kinney T.N., Zhang W., Gibney, T., Pani, A.M., Stern, M.J., Matus, D.Q. Understanding the Quiescence to Proliferative Switch in vivo. 2021 ASCB Annual Meeting |